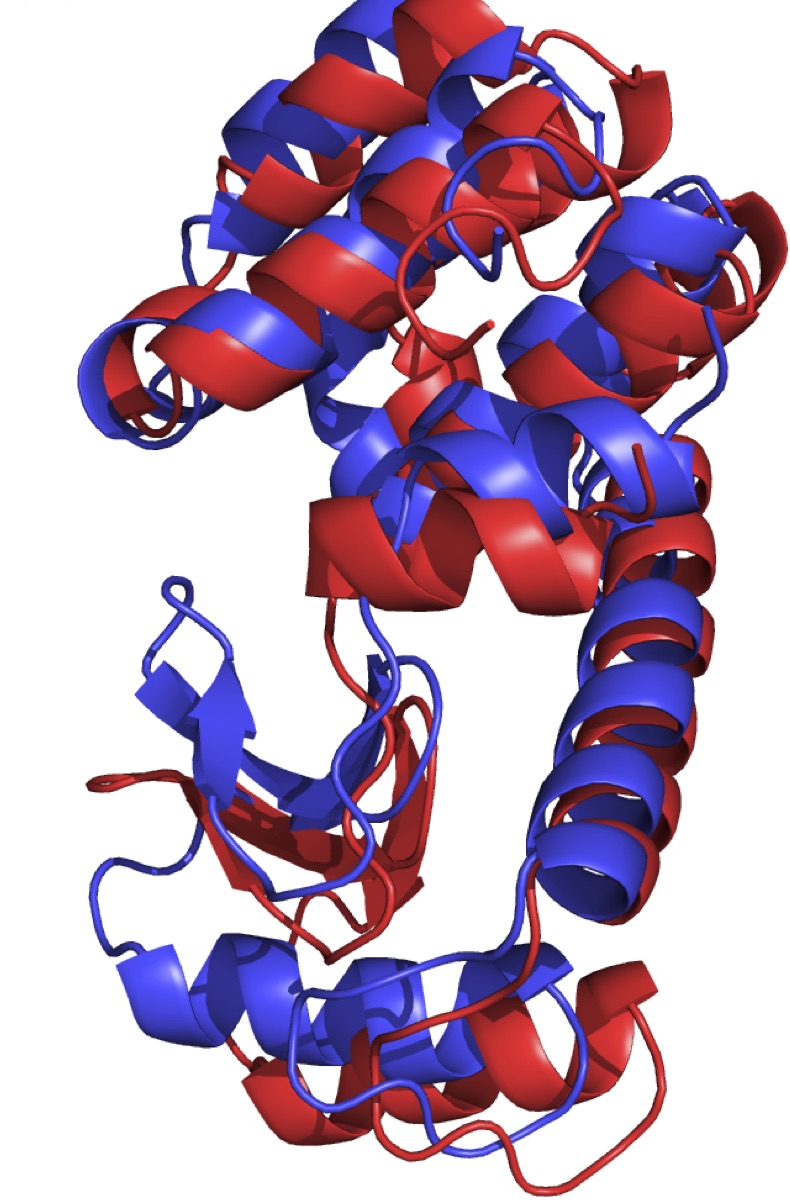
T4 lysozyme is an enzyme is used by enterobacteria phage T4 to destroy bacterial cell walls by catalyzing the cleavage of glycosidic bonds. The interaction of T4L with the substrate involves opening and closing of the protein structure, mediated by prominent hinge-bending motion of the two domains. While it has been extensively modelled, the mechanism of this conformational change is not well understood in terms of reaction coordinates [1]. In this project we will use transition path sampling to investigate the mechanism. We will collect and analyse an ensemble of reactive dynamics pathways, that can be scrutinized for important collective variables, to arrive at a prospect reaction coordinate. A proposed four state model will be tested.
[1] Ernst, M., Wolf, S., & Stock, G. (2017). Identification and Validation of Reaction Coordinates Describing Protein Functional Motion: Hierarchical Dynamics of T4 Lysozyme. Journal of Chemical Theory and Computation, 13(10), 5076–5088. https://doi.org/10.1021/acs.jctc.7b00571