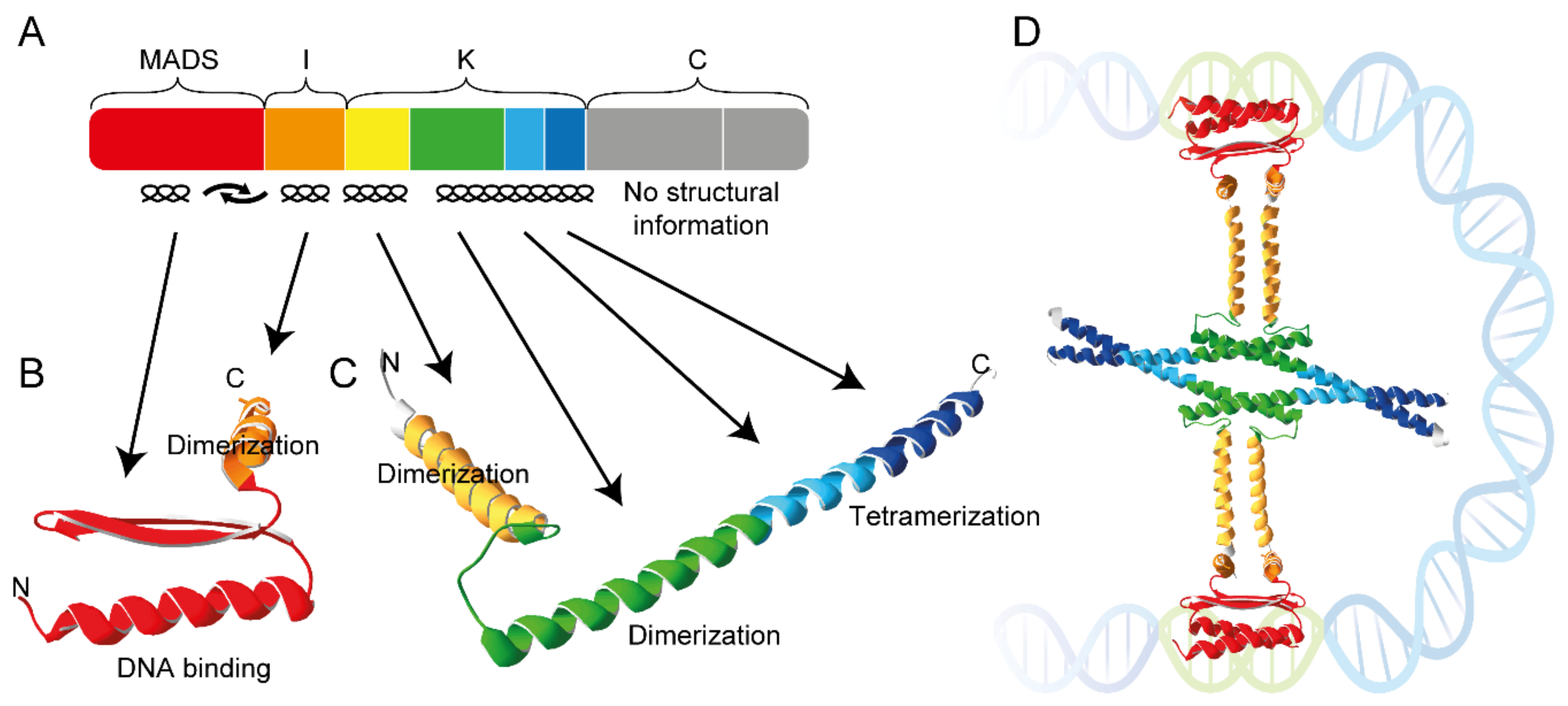
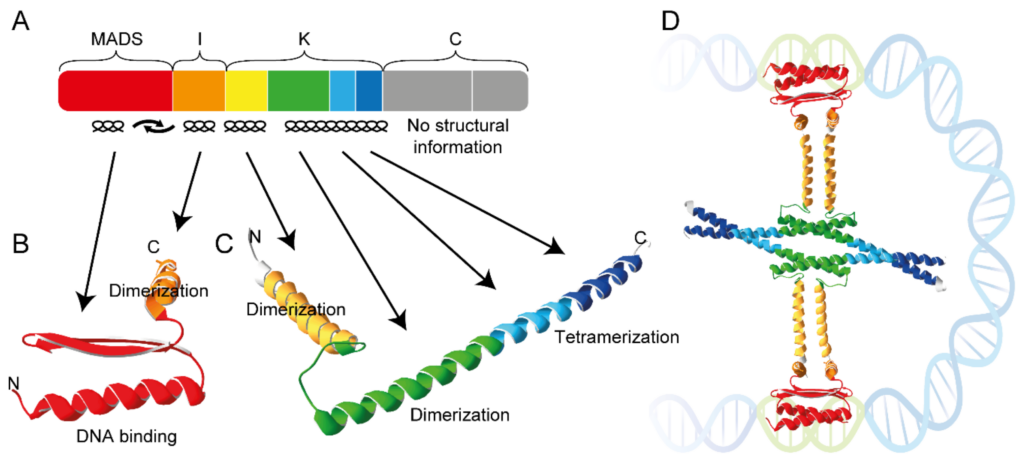
MADS domain transcription factors are involved in regulating a variety of processes in plants, ranging from root formation to floral organ development and timing of the floral transition. The MADS protein domain family has many members in plants and any given plant may contain 50 or more family members [Smaczniak2017]. MADS domain proteins form various homo- and hetero- dimers and tetramers. Specifically, tetramerization is mediated by coiled-coil interactions between one of the subdomains of the proteins, the so-called K-domain. The aim of this project is to characterise and quantify the interaction between K-domains from different MADS domain transcription factors using molecular simulation.
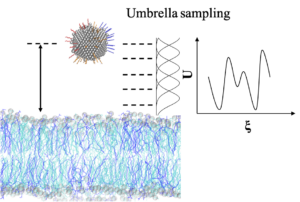
To become familiar with the approach as outlined in Ref. [vanHeesch2023], microsecond long MD simulations will be conducted starting with a protein-protein complex, to map the interactions between the two proteins. Then, the free energy difference of dissociation will be computed using steered molecular dynamics. The procedure will be repeated for various protein-protein complexes involving K-domains from Arabidopsis MADS proteins for which structures are available [PDB 4OX0], this procedure will be preceded by short MD simulations of the K- domain. Structural models for additional Arabidopsis MADS proteins will be obtained with AlphaFold2 [Jumper2021].
The project will result in a list of interaction free energies between different MADS proteins which can be used in various data science and machine learning approaches.
The project will result in a list of interaction free energies between MADS proteins and DNA, which can be used in various data science and machine learning approaches.
This project is in collaboration with prof. dr. Aalt-Jan van Dijk at the Swammerdam Institute for Life Sciences