Bachelor/Master Project
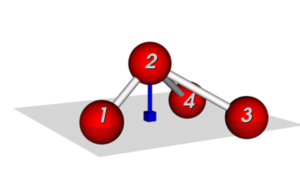
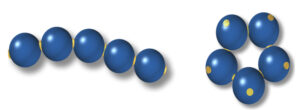
Modeling out-of-equilibrium colloidal patchy particle architectures
The student will conduct large scale simulations of patchy particles, using transition path sampling, in order to understand rare transitions in active colloidal systems.